|
||||||
|
DNA Input as Fuel | Biological Nanocomputer | Turing Machine-Like Model
|
||||
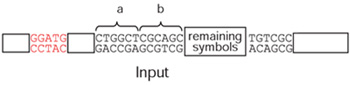
One of the paramount goals of nanotechnology is the creation of nanocomputers. While nanocomputers could have numerous applications, the one that stirs the imagination the most is the launching of nanocomputers inside the human body on an in vivo mission to identify malfunctions and fix them. Will such a "fantastic voyage" ever be possible? And if so, what steps can we take to expedite embarkation? We believe this vision is realizable. Molecular machines inside the living cell already posses a full repertoire of operations required to implement a universal computer (such as a Turing machine) and science's understanding of the cell's molecular machinery is improving by the day. But, as the ability to create a molecular machine "from scratch" may be several decades away, one approach is to hunt nature's "mines" for existing molecular machines that can be stitched together and coerced to compute, even if in a somewhat cumbersome way. We recently reported an effort to construct such a simple computer (Abstract). The computer's input, output, and "software" are made up of double-strand DNA molecules. Two naturally occurring enzymes that manipulate DNA form the "hardware." These are FokI, an enzyme that cuts DNA, and Ligase, an enzyme that seals two DNA molecules together. When mixed in solution, the input molecule is processed by software and hardware molecules to create the output molecule, which contains the result of the computation. This simple mathematical computing machine is known as a finite automaton. By choosing different software molecules to be mixed in solution, this nanocomputer can be programmed to perform several simple tasks. For instance, it can detect whether, in an input molecule encoding a list of 0's and 1's, there are an even number of 1's. The software molecules can be used to create a total of 765 software programs. A number of these programs were tested in the lab, including the "even 1's checker" mentioned above, as well as programs that check whether a list of 0's and 1's has all the 0's before all the 1's, whether it has at least (or at most) one 0, and whether it both starts with a 0 and ends with a 1. How can DNA strands to contain the symbols 0 and 1? DNA strands are usually depicted as a scroll of recurring "letters," in varied combinations, that represent DNA's constituents (four chemical bases). The team decided that the letter pattern "CTGGCT" in the input molecule would signify "1" (a in the diagram below) and "CGCACG" would signify "0" (b in the diagram).
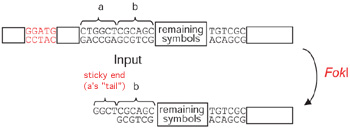
The input molecule, when mixed with hardware and software molecules, also two "states". When the hardware molecule FokI, recognizing a symbol, "cuts" DNA, it leaves it with one strand longer than the other, resulting in a single-strand overhang called a "sticky end" (see diagram below). Since FokI makes its incision at the site of the symbol, the "sticky end" is what remains of the symbol. FokI may leave the symbol's "head" or "tail" attached. These are the two possible "states."
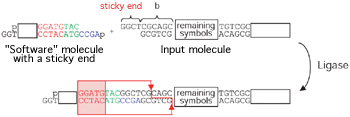
Two molecules with complementary sticky ends can temporarily stick to each other (a process known as hybridization). In each processing step the input molecule hybridizes with a software molecule that has a complementary sticky end, allowing the hardware molecule Ligase to seal them together using two ATP molecules as energy (see diagram below).
Then comes Fok-I, and cleaves the input molecule again, in a location determined by the software molecule. Thus a sticky end is again exposed, encoding the next input symbol and the next state of the computation. Once the last input symbol is processed, a sticky end encoding the final state of the computation is exposed and detected, again by hybridization and ligation, by one of two "output display" molecules. The resulting molecule, which reports the output of the computation, is made visible to the human eye in a process known as gel electrophoresis. The automaton is so small that 1012 automata sharing the same software run independently and in parallel on inputs (which could in principle be distinct) in 120 ml solution at room temperature. Their combined rate is 109 transitions per second, their transition fidelity is greater than 99.8%, and together they consume less than a billionth of one Watt. As science's understanding of the cell's molecular machinery is rapidly improving, there is no reason to expect an insurmountable obstacle to transforming this dream into a reality. Eventually, one will be able to harness nature's building blocks and techniques to construct a molecular machine to specification. |
||||