Molecular computer fueled by its DNA input
The field of experimental molecular computing, almost a decade old by now, continues to throw new light onto many aspects of computation. Our recent research mainly deals with the aspect of energy consumption by a computer. We were able to construct a molecular computer whose sole energy source is its input, a combination unthinkable of in the realm of electronic computers. This energy is extracted as the input data molecule is destroyed during computation. As a side-effect we were able to modify our previous molecular computer so that it does not consume software molecules during computation: the only component that changes is the input, while the hardware and the software molecules remain unchanged.
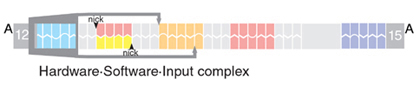
The computation capabilities of the computer are similar to that of the previous one. The computer is a special case of a Turing machine, a two-state, two-symbol finite automaton and as such it is able to answer simple questions about binary strings, such as “Does a binary string of a’s and b’s contain an even number of a’s?” or “Is the length of the string even or odd?” A sample computation on an input string abba, which gives an answer to the question about the even a’s is presented below:

The combination of the initial automaton state and input string (S0-abba) as well as the combinations of the intermediate state and the remaining input symbols are all ‘configurations’ of an automaton. A particular set of the transition rules defines a question that the automaton can answer, and therefore comprises its program. The computation is a succession of configurations, which are generated by applying in turn a suitable transition rule from the program set to a configuration. In our automaton, each configuration, as well as each transition rule, are encoded by a single molecule.
The computation starts when one of the transition rules is applied to a molecule that encodes an initial configuration – an initial state and an input string. The input string is encoded in a DNA molecule by deliberate placing side-by-side of pre-designed stretches of five nucleotides long, interrupted by spaces three nucleotides long. The initial state and the intermediate states are encoded by a modification of the leftmost symbol of an input string: The double-stranded piece of DNA encoding a symbol is cleaved and becomes partially single-stranded. While double-stranded DNA is inert towards other DNA molecules, single-stranded DNA readily interacts with other single-stranded DNA molecules in a complementary fashion. To encode different states, different portions of the symbol are exposed to the environment: the leftmost four nucleotides represent state S1 while the rightmost four represent the state S0:
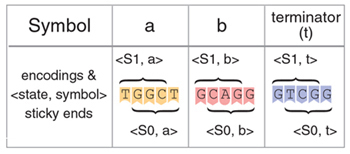
Each single-stranded stretch, known sometimes as “sticky ends”, uniquely represents both a current state and the leftmost (current) symbol of an automaton configuration. We term them “<state, symbol> sticky ends”.
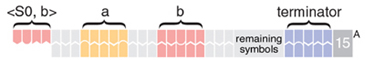
The complete encoding of a configuration in a molecule is formed by placing a modified leftmost input symbol followed by double-stranded stretches representing the remaining symbols. For example, the molecule below represents an input in a configuration S0-bab.
The transitions between intermediate configurations towards the final output configuration is performed by the ‘hardware’ and the ‘software’ segments of the computer. The hardware is an SII type restriction endonuclease FokI, an enzyme that is capable to recognize well-defined segment of double-stranded DNA, termed ‘recognition site’ and cleave both strands at constant, but different, distances from this site. The cleavage of DNA is a spontaneous process, driven by the energy released upon the destruction of the DNA backbone.

The software molecules represent the transition rules. Each molecule contains a binding site for the FokI enzyme and a single-stranded stretch of DNA that is complementary to the <state, symbol> sticky end of the input. The binding site and the sticky end are separated by a spacer of variable length, which determines the location of the cleavage sites within a next symbol of the input molecule and therefore defines the next state of the automaton:
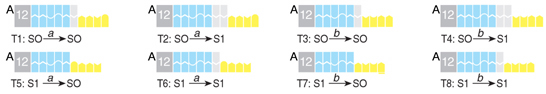
We found that the noncovalent interaction between the sticky ends of the software and the input molecule is enough to direct the FokI to cleave at the required location. Unlike in the previous computer, the cleavage did not require covalent ligation between the molecules. Therefore, the driving force behind the transition is the heat dissipation and entropy increase that accompany DNA cleavage, and not ATP hydrolysis as in the previous computer. The mechanism of computation is outlined in the following scheme:
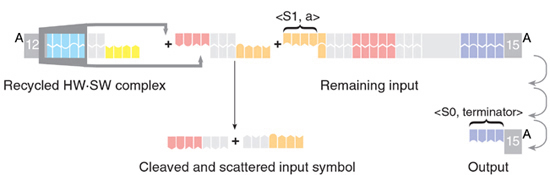
1) A complex consiting of a software molecule and a hardware molecule approaches the input:
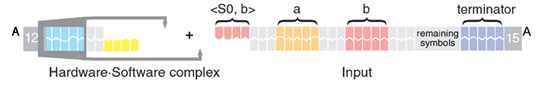
2) The noncovalent hybridization between the sticky ends of the software and the input position the cleavage sites of the FokI enzyme at the required locations within the next symbol:
3) The hardware enzyme processes the input, exposing the next <state, symbol> tag and therefore transferring the computation into its next configuration. At the same time, current symbol is cleaved off the input molecule, re-dissociates from the software molecule and turns into garbage. The software and the hardware molecules are recycled and may participate in future rounds of the computation. Eventually, an output is formed.
To confirm that the computation proceeds according to our model, we analyze the output directly using radioactive labelling of the input. We know the expect lengths of the “tails” representing different outputs and we follow their formation by a denaturing gel electrophoresis.
The new computer has greatly improved performance characteristics compared to our previous one. The fastest computation optimized for a single input molecule required only 20 seconds per step per molecule, about 50-fold improvement. Under optimized parallel performance, the cumulative number of operations was 6.646x1010 operations per second per ml, an ~8000-fold improvement over the previous system. Heat dissipation due to input destruction is about 5.3x10-9 W/ l.
l.
PNAS advanced online publication

